Plink_Assoc_Viewer
About this program
This program allows the output from a PLINK association analysis to be visualised.
Each analysis calculates the 'chi-square value (1df'), 'Asymptotic p-value' and
'Estimated odds ratio' results for each SNP. These results are viewed one chromosome
at a time by selecting the analysis type from the drop down list in the centre of
the window and the chromosome from the drop down list in the lower left corner of
the window. The 'chi-square value (1df') and 'Estimated odds ratio' are displayed
on a linear axis, while 'Asymptotic p-value' values are shown as the negative Log
10 of the value.
Data for a region can be exported as either a tab delimited text file or a html
web page. To export data place the cursor at one side of the region and with the mouse
button pressed move the cursor to the other end of the region. Then press the
Save button and select the correct file extension and enter
the name of the file to save the data too.
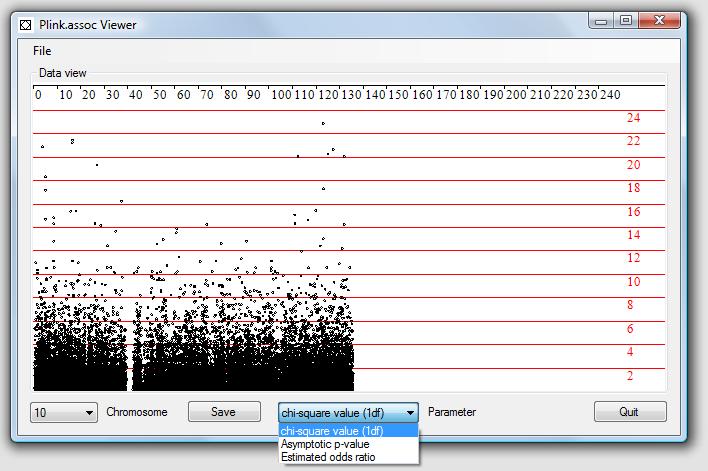
Figure 1
The data in a plink.assoc file is arranged as 10 fixed width
columns as described in Table 1, the second row states the width of each column,
while the third row shows example data. If you analyse the data with the --ci X
arguement the file will contain and extra 3 columns, these will be ignored by this
program.
| Chromosome |
SNP |
Position |
Minor allele |
Minor allele frequency in affecteds |
Minor allele frequency in unaffecteds |
Major allele |
chi2 value |
Asymptotic p-value |
Estimated odds ratio |
| 4 |
13 |
11 |
5 |
9 |
9 |
5 |
13 |
13 |
13 |
|
10 |
RS1530116 |
122657012 |
A |
0.1083 |
0.3889 |
C |
22.96 |
0.000001656 |
0.1909 |
 Plink_assoc_Viewer can be downloaded from here
Plink_assoc_Viewer can be downloaded from here
Note: this program requires the .NET framework version 2.0 to be installed
(available through Windows Update or
here).
- Plink details
- Plink was developed by Shaun Purcell at the Center for Human Genetic Research,
Massachusetts General Hospital and has it own web page
here
- PLINK citation
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D,
Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. (2007).
PLINK: a tool set for whole-genome association and population-based linkage analyses.
Am J Hum Genet. 81(3):559-75.
|